Smart Health and Wellbeing
Machine Learning Models for Blood Glucose Prediction in Diabetes Management
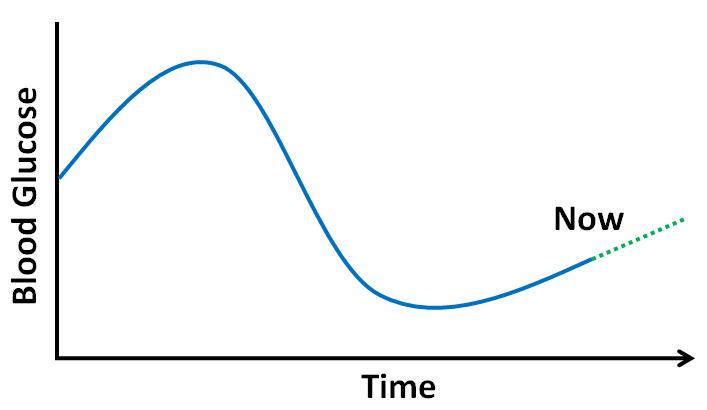 Good blood glucose control is essential for patients with diabetes to avoid serious complications
of the disease. However, achieving and maintaining good blood glucose control is difficult.
Patients are highly individual in their responses to treatment and to life events that affect
blood glucose levels. Large volumes of blood glucose data may be collected automatically,
but automated analysis is lacking. Patients do not always know when problems are impending;
problems occurring while patients are asleep are especially dangerous. Machine learning
models that predict blood glucose levels would enable or facilitate new applications of
direct benefit to patients, including: alerts to immediately notify patients of impending
problems; decision support systems recommending actions to prevent problems; and educational
simulations showing the effects of different treatment choices or lifestyle options on blood
glucose levels.
Good blood glucose control is essential for patients with diabetes to avoid serious complications
of the disease. However, achieving and maintaining good blood glucose control is difficult.
Patients are highly individual in their responses to treatment and to life events that affect
blood glucose levels. Large volumes of blood glucose data may be collected automatically,
but automated analysis is lacking. Patients do not always know when problems are impending;
problems occurring while patients are asleep are especially dangerous. Machine learning
models that predict blood glucose levels would enable or facilitate new applications of
direct benefit to patients, including: alerts to immediately notify patients of impending
problems; decision support systems recommending actions to prevent problems; and educational
simulations showing the effects of different treatment choices or lifestyle options on blood
glucose levels.
The task of blood glucose prediction is approached as a time series forecasting problem.
Blood glucose is predicted based on a patient's prior blood glucose levels, insulin data, meal data,
exercise data, sleep patterns and work schedules. Batch and incremental time series regression models,
including support vector machines and neural networks, are being investigated. To account for
individual patient differences, separate models are trained for each patient. Additionally,
transfer learning may enable data from multiple patients to aid in building models for patients
with limited historical data. Models are sought that are robust in the face of imperfect data,
including missing life events, inaccurately recorded life events, and noisy glucose sensors. This was the original founding project of the SmartHealth Lab at Ohio University, which is dedicated to promote interdisciplinary work in smart health and wellbeing.
Our work aims to improve the overall health and wellbeing of patients with diabetes. Toward this aim, we release the code for our SVR+Physio model for blood glucose prediction. This code contains the implementation of a physiological model with Kalman filtering, which is used to provide features to a Support Vector Regression model.
This material is based upon work supported by the National Science Foundation
under Grant Number 1117489. Any opinions, findings, and conclusions or
recommendations expressed in this material are those of the author(s) and
do not necessarily reflect the views of the National Science Foundation.
 Good blood glucose control is essential for patients with diabetes to avoid serious complications
of the disease. However, achieving and maintaining good blood glucose control is difficult.
Patients are highly individual in their responses to treatment and to life events that affect
blood glucose levels. Large volumes of blood glucose data may be collected automatically,
but automated analysis is lacking. Patients do not always know when problems are impending;
problems occurring while patients are asleep are especially dangerous. Machine learning
models that predict blood glucose levels would enable or facilitate new applications of
direct benefit to patients, including: alerts to immediately notify patients of impending
problems; decision support systems recommending actions to prevent problems; and educational
simulations showing the effects of different treatment choices or lifestyle options on blood
glucose levels.
Good blood glucose control is essential for patients with diabetes to avoid serious complications
of the disease. However, achieving and maintaining good blood glucose control is difficult.
Patients are highly individual in their responses to treatment and to life events that affect
blood glucose levels. Large volumes of blood glucose data may be collected automatically,
but automated analysis is lacking. Patients do not always know when problems are impending;
problems occurring while patients are asleep are especially dangerous. Machine learning
models that predict blood glucose levels would enable or facilitate new applications of
direct benefit to patients, including: alerts to immediately notify patients of impending
problems; decision support systems recommending actions to prevent problems; and educational
simulations showing the effects of different treatment choices or lifestyle options on blood
glucose levels.


